
Mattias E. Jonasson
@mattejonasson
Enhancer synergy in stem cells | PhD student in the Bücker lab
ID: 1560662173529133057
19-08-2022 16:17:39
11 Tweet
35 Followers
83 Following

Happy sharing insights from the lab of Claudio Cantù at the Abcam Helmholtz Munich | @HelmholtzMunich summer school on chromatin biology - great experience!


Our paper on the architecture of the #chikungunya virus replication organelle is out eLife - the journal. Congrats to our PhD student Tim Laurent and all other authors! A 🧵on the main findings. (1/8) #teamtomo #cryoEM elifesciences.org/articles/83042



Our CUT&RUN-LoV-U protocol is now online Development doi.org/10.1242/dev.20… Please use it to map the genome-wide binding pattern of non-DNA binders FYI, it works well also with all types of targets We'd LoV ❤️ to know how this protocol works in your hands - please feedback!
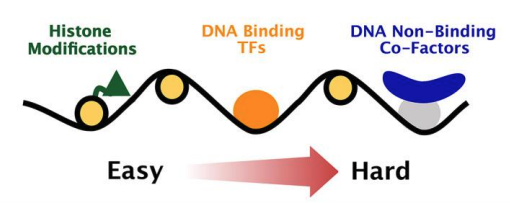

A new CUT&RUN low volume-urea (LoV-U) protocol optimized for transcriptional co-factors uncovers Wnt/β-catenin tissue-specific genomic targets An #OpenAccess Techniques and Resources Article from Gianluca Zambanini, Claudio Cantù, et al Linköpings universitet journals.biologists.com/dev/article/14…


Two great new methods out today in Nature Biotechnology on multi-target chromatin profiling using nanobody-tethered Tn5 transposases: 1) NTT-seq from Raimondi Ivan and Rahul Satija... nature.com/articles/s4158… 1/3

The Time-Resolved Genomic Impact of #WNT/β-catenin signaling Cell Systems Cell Press cell.com/cell-systems/f… We show -β-catenin repositions over TIME ⏰ -How this impacts global transcription⬆️⬇️ -Preceding and following chromatin dynamics🧬 Efforts driven by Pierfrancesco Pagella
