
Igor Dolgalev
@bioigor
Assistant Professor @NYUGSOM_PMED @nyugrossman
ID: 870030789483868161
https://igor.pub 31-05-2017 21:34:39
948 Tweet
398 Followers
294 Following




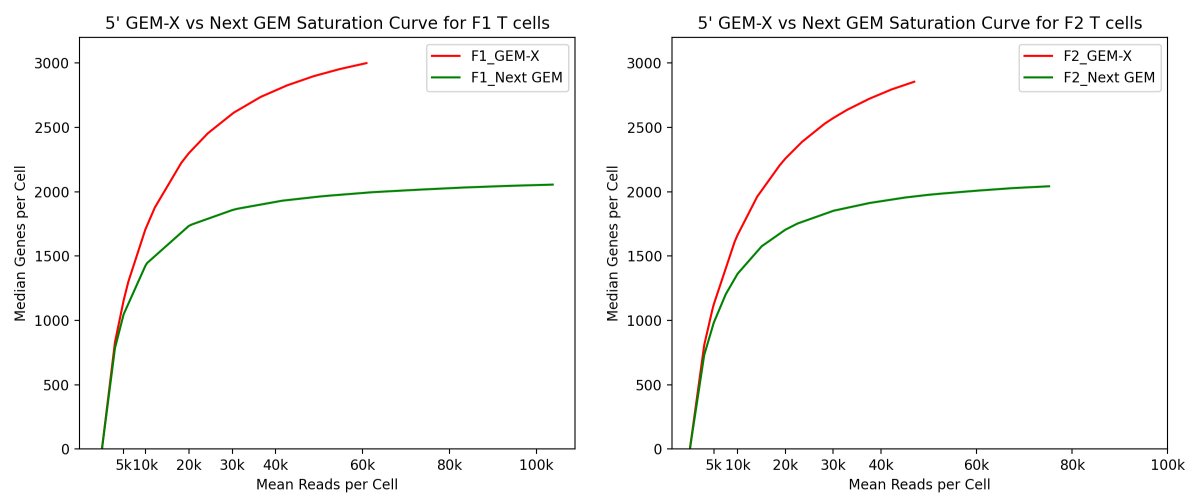
10x Genomics released new single cell GEM-X assays in March, and we were quick to try it out. FHIL benchmarked 5’ GEM-X against 5’ Next GEM v2 chemistry with sorted T-cells from healthy donor PBMCs and targeted the recommended max for each assay, 20k and 10k respectively.(1/8)

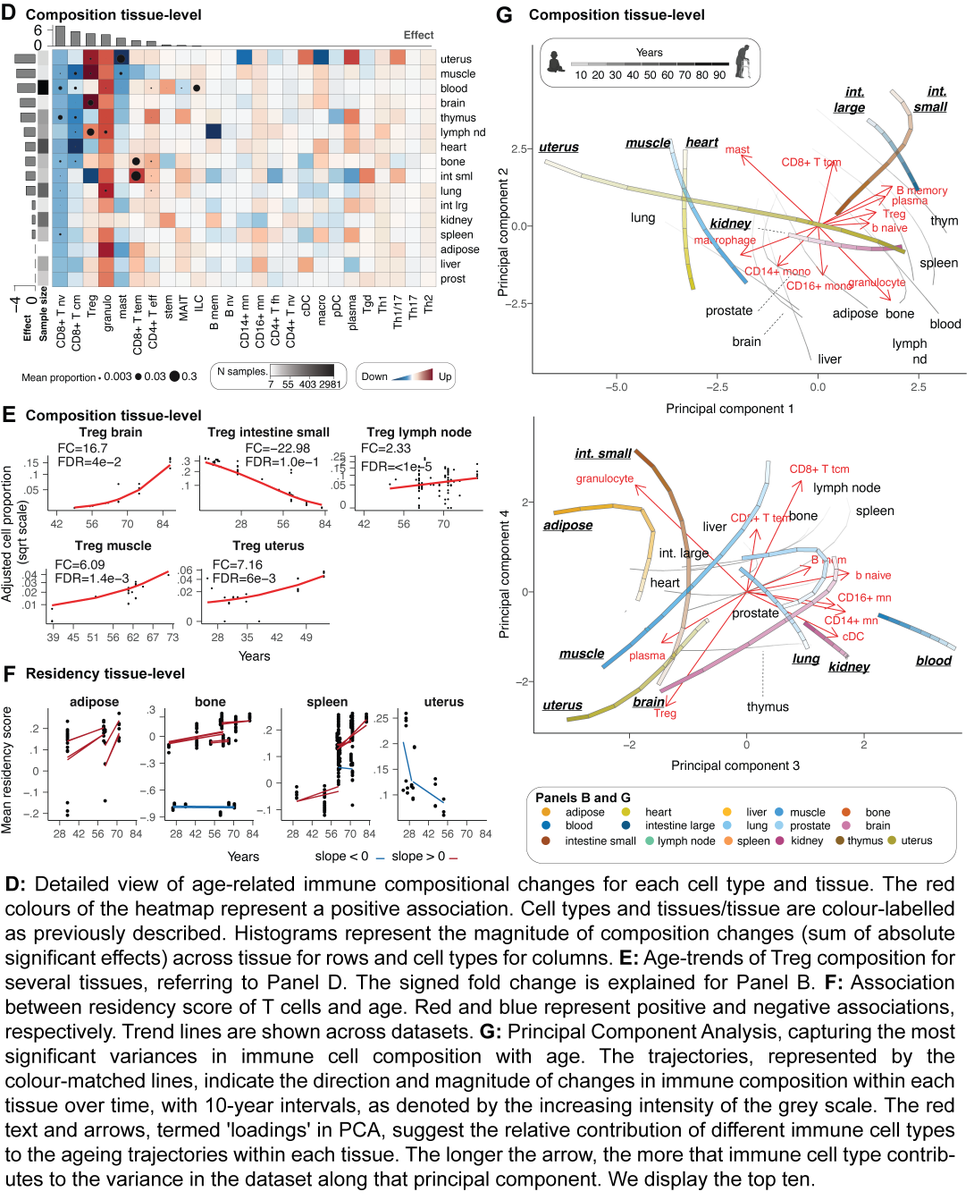
🌎👵⚤ Excited to share Multi-organ map of the human immune system across age, sex and ethnicity From 29M cells and 12K samples from Human Cell Atlas, tissue-specific #Ageing programs, sexual #dimorphism, #Ethnical diversity in #Immunotherapy targets biorxiv.org/content/10.110…

I'm happy to share that out paper on Phantasus, a web-application for visual and interactive gene expression analysis, got out in eLife - the journal elifesciences.org/articles/85722 Here, I'm using it to analyze a random GEO dataset, including basic QC to filter outliers, in under 100 seconds! 1/n


Systematic comparison of sequencing-based spatial transcriptomic methods in Nature Methods In summary: “- Stereo-seq, Slide-tag, Visium shows the better capture efficiency with raw sequencing depth - Slide-seq V2, Visium (probe), DynaSpatial gives the better capture efficiency



Are you missing cells and cell types in your single-cell multiome experiments 10x Genomics? Check out EmptyDropsMultiome MarioniLab


