
TomDendooven
@tdendooven
ID: 997923431902580736
19-05-2018 19:34:21
31 Tweet
191 Followers
160 Following



This is exactly what #teamtomo needs. Nice work Alister Burt et al. 💪💪 Next step is to add fiducial-less tilt-series alignment to the front and deep learning picking and segmentation to the back. biorxiv.org/content/10.110…

Hey #teamtomo - our lab (Barford Group MRC Laboratory of Molecular Biology) are looking for a postdoc to join us in our efforts to understand kinetochore structure and chromosome segregation! Retweets appreciated, DMs very much open for informal enquiries about life here mrc.tal.net/vx/mobile-0/ap…


#CryoEM structure of the complete inner kinetochore of the budding yeast point centromere. An amazing set of structures from the Barford lab MRC Laboratory of Molecular Biology, including results from our newest tool by Johannes Schwab to handle structural flexibility. 🥳 biorxiv.org/content/10.110…




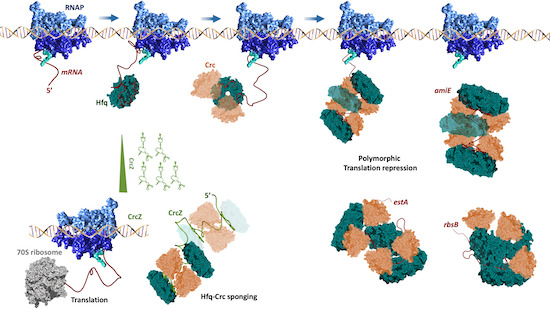
How does bacterial RNA #chaperone #Hfq regulate numerous different mRNAs and processes? #CryoEM by TomDendooven Ben Luisi Cambridge University and collaborators shows assembly into distinct polymorphic #ribonucleoprotein complexes as basis embopress.org/doi/10.15252/e…



Happy to see one of the last pieces of my PhD work published! Very lucky to have awesome TomDendooven as a partner in crime Also many thanks to Alister Burt, Dom, John, David and Rappsilber laboratory

When I joined David Barford's group MRC Laboratory of Molecular Biology it was with the goal of using my cryo-ET and computational skills to help them understand funky biology. I joined TomDendooven and Stanislau Yatskevich's efforts to average the native gamma tubulin ring complex (γTuRC) from tomograms of SPBs

Over 34 years ago, MRC-LMB Cell Biology's John Kilmartin first purified yeast spindle bodies with their attached spindle microtubules. Still going strong he has now collaborated with David Barford's lab to reveal how gamma tubulin nucleates microtubules.

New #CryoET structure of the native γ-tubulin ring complex gives insight into microtubule nucleation. Read more about the study led by TomDendooven & Stanislau Yatskevich in David Barford’s group at the LMB here: www2.mrc-lmb.cam.ac.uk/structure-of-%… #LMBResearch LMB PhD Programme Rappsilber laboratory Alister Burt

