anna brogan
@apbrogan
PhD student into bacteria & coffee. Rudner lab @harvardmed. @penn_state alum.
ID: 1127335867981365249
11-05-2019 22:13:10
48 Tweet
181 Followers
204 Following


So excited to share this work from the Walker Lab, now online Nature Microbiology (nature.com/articles/s4156…), in which we uncover a general strategy for bacterial cell envelope polymer acylation using teichoic acid ᴅ-alanylation as a paradigm! A 🧵 below for anyone interested:












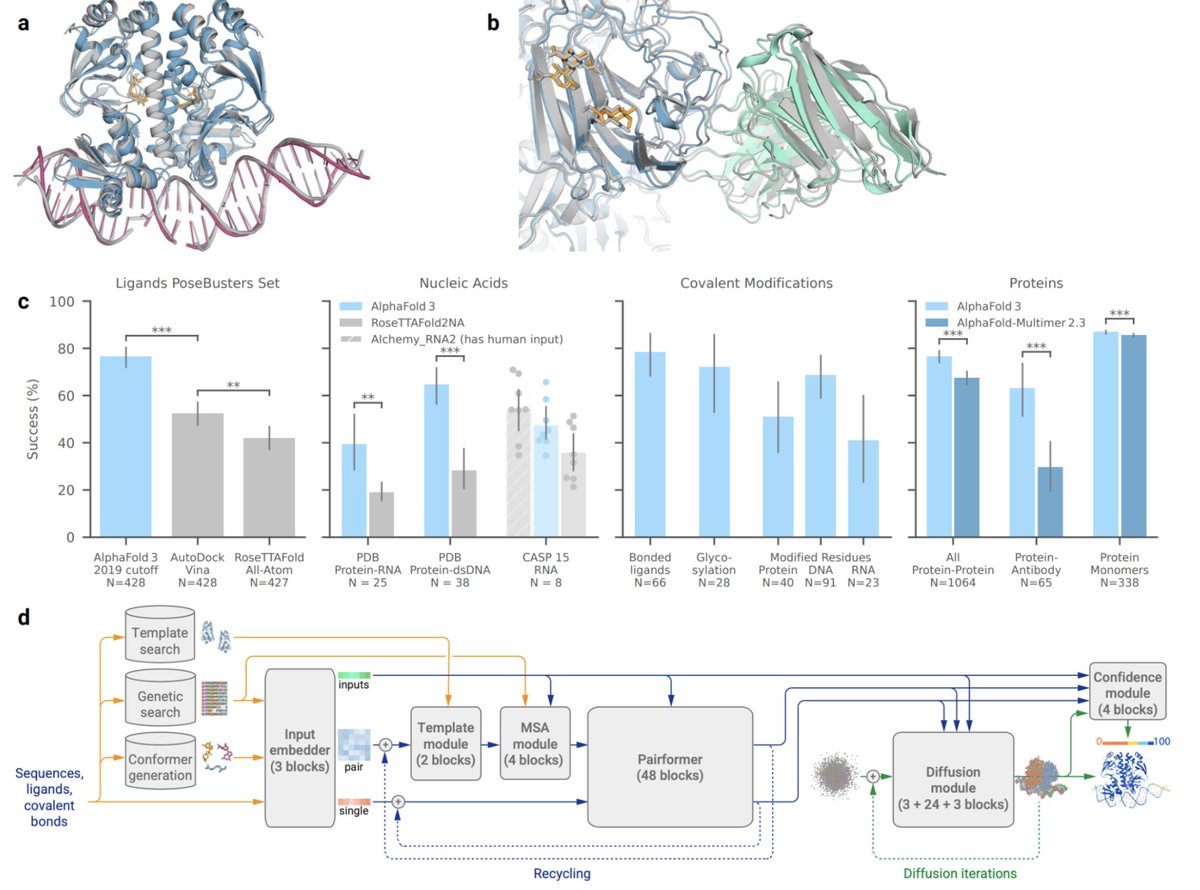
Super excited to be releasing AlphaFold 3 today, developed by Isomorphic Labs and Google DeepMind: our next generation AI model for predicting the biomolecular structures and interactions of proteins, DNA, RNA, small molecules, and more: bit.ly/44yfaCw 1/


another great year for Boston Bacterial Meeting (BBM)! huge shoutout to all of the presenters, the OC, and to Laurent Dubois and Molly Sargen for running the whole show (& for incredible crisis management when BBM went dark)


Excited to share that our new preprint is out! We used #AlphaFold multimer to screen 7000+ nuclear proteins to find new nucleosome acidic patch binders. Great collaboration between Johannes Walter and Lucas Farnung. biorxiv.org/content/10.110…