Fengchao Yu
@fcyucn
Research Investigator at U of M. Interested in proteomics, etc. Developer of FragPipe, MSFragger, IonQuant, etc.
ID: 1388236398
https://fcyu.github.io/ 29-04-2013 00:24:23
402 Tweet
780 Followers
739 Following







Very interesting bioRxiv by the group of Alexey Nesvizhskii and Fengchao Yu. They developed the tool diaTracer for fully integrated, library-free analysis of diaPASEF data from a Bruker timsTOF in FragPipe. Looking forward to exploring this on our data. biorxiv.org/content/10.110…


A key piece of this study was the new PAL localization mode that is built into the new #FragPipe V22 release thanks to Fengchao Yu and Alexey Nesvizhskii


Fengchao Yu Alexey Nesvizhskii FragPipe now reports the hyperscores for each amino acid in a peptide sequence based on both shifted (PAL modified) and unshifted ions, which allows the label to be localized to either a single amino acid or within a region of the peptide sequence, depending on sequence coverage



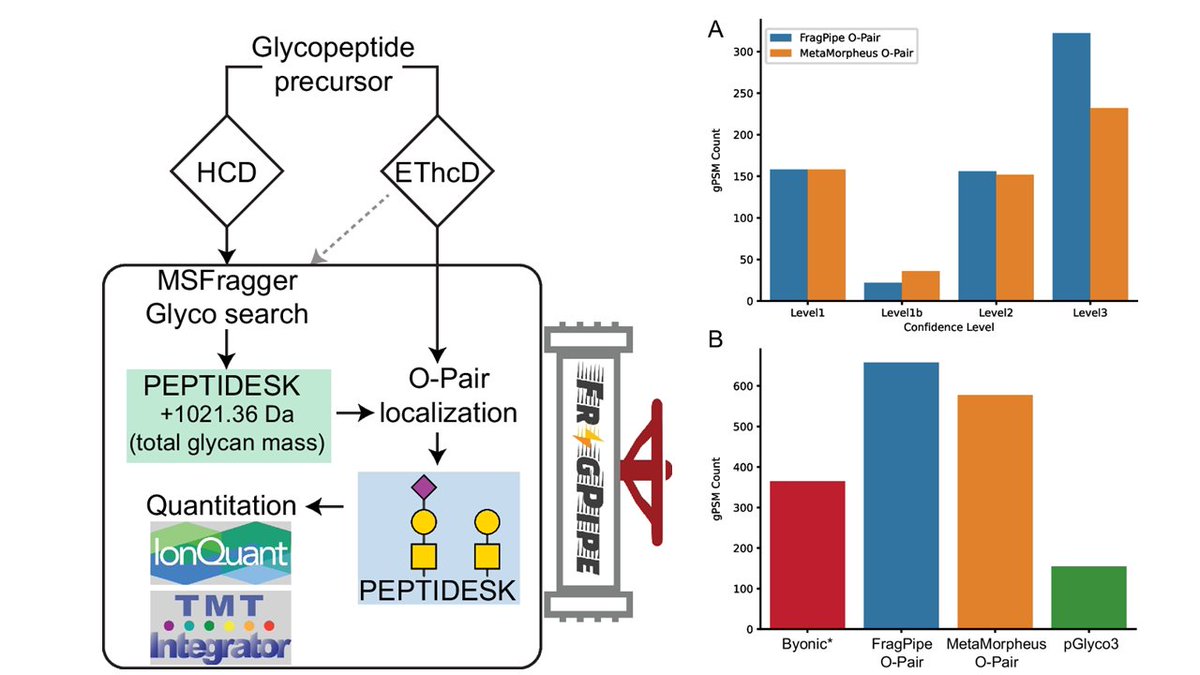
Led by Dan Polasky and in collaboration with MetaMorpheus @metamorpheus.bsky.social lab, we improved #FragPipe for O-glycoproteomics analysis. It provides fast (#MSFragger-Glyco), proteome-wide, quantitative results (LFQ,TMT), and incorporates O-Pair for glycosite localization! link.springer.com/article/10.100…

I am enjoying my visit to Madison to give a lecture and a short #FragPipe tutorial at the North American MS Summer School ncqbcs.com/resources/trai…. At the some time, Fengchao Fengchao Yu is flying to San Francisco to do the same at the UCSF proteomics course. wiitalab.ucsf.edu/events/biologi….


Fun running our 1st hands-on “biological #proteomics for beginners” workshop @ucsf w Dr. Sanjeeva Srivastava. packed house for scientists across the Bay Area. Thanks to Fengchao Yu for coming from Ann Arbor to lead our analysis tutorial in Fragpipe and Rob Moritz for keynote lecture!







Our #FragPipe-Analyst manuscript is out! pubs.acs.org/doi/10.1021/ac…. Load FragPipe's TMT/ LFQ/DIA quant tables (peptide or protein quant) and get QC, Limma, PCA, volcano plots, heatmaps, do GO/pathway enrichment analysis, all with just a few clicks! Tutorials:fragpipe-analyst-doc.nesvilab.org