Jeroen Gilis
@gilisjeroen
PhD student, developing statistical software for single-cell transcriptomics analysis, Ghent university
ID: 1139806244737310720
https://jgilis.github.io/ 15-06-2019 08:06:00
389 Tweet
255 Followers
529 Following

Alternative splicing as a source of phenotypic diversity go.nature.com/3yW3Q3V #Review by Charlotte Wright, Christopher W. J. Smith & Chris Jiggins Wellcome Sanger Institute Darwin Tree of Life Dept of Zoology Cambridge Biochemistry RNA labs



Interested to know how #reprocessing and reusing existing #proteomics #data helps uncover the unknown? Let's have a beer and talk about this at DataBeers Brussels this Thursday. Details below! See you soon! #bigdata #humanproteome

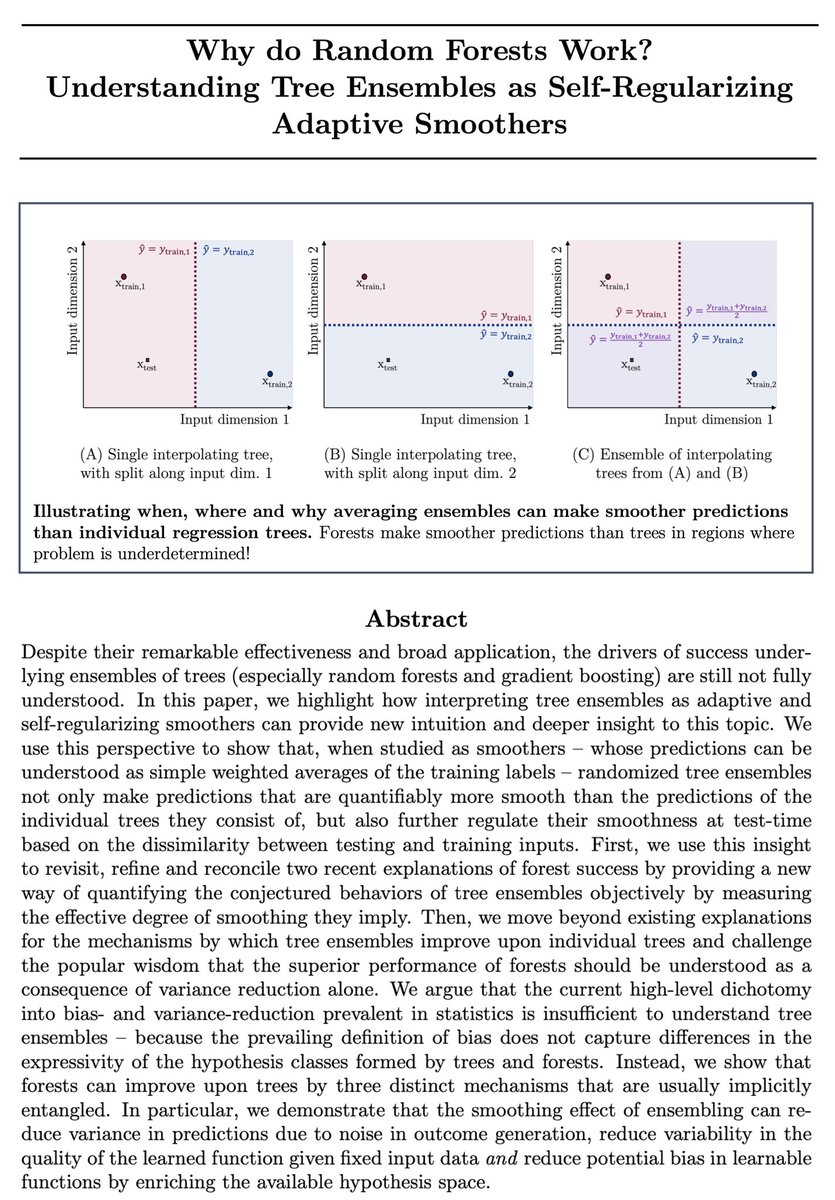
Why do Random Forests perform so well off-the-shelf & appear essentially immune to overfitting?!? I’ve found the text-book answer “it’s just variance reduction 🤷🏼♀️” to be a bit too unspecific, so in our new pre-print arxiv.org/abs/2402.01502, Alan Jeffares & I investigate..🕵🏼♀️ 1/n

.Hector RdB, @koenvdberge_be, Kelly Street & Sandrine Dudoit present condiments for the inference and downstream interpretation of cell trajectories across multiple conditions #BiotechNatureComms nature.com/articles/s4146…





I am deeply honored and humbled to be recipient of an advanced ERC award #ERCAdG to spearhead research on Assumption-lean (Causal) Modeling and Estimation. Ghent Data Analysis and Statistical Science @UGent_fwe Ghent University Research @UGent European Research Council (ERC) 1/2

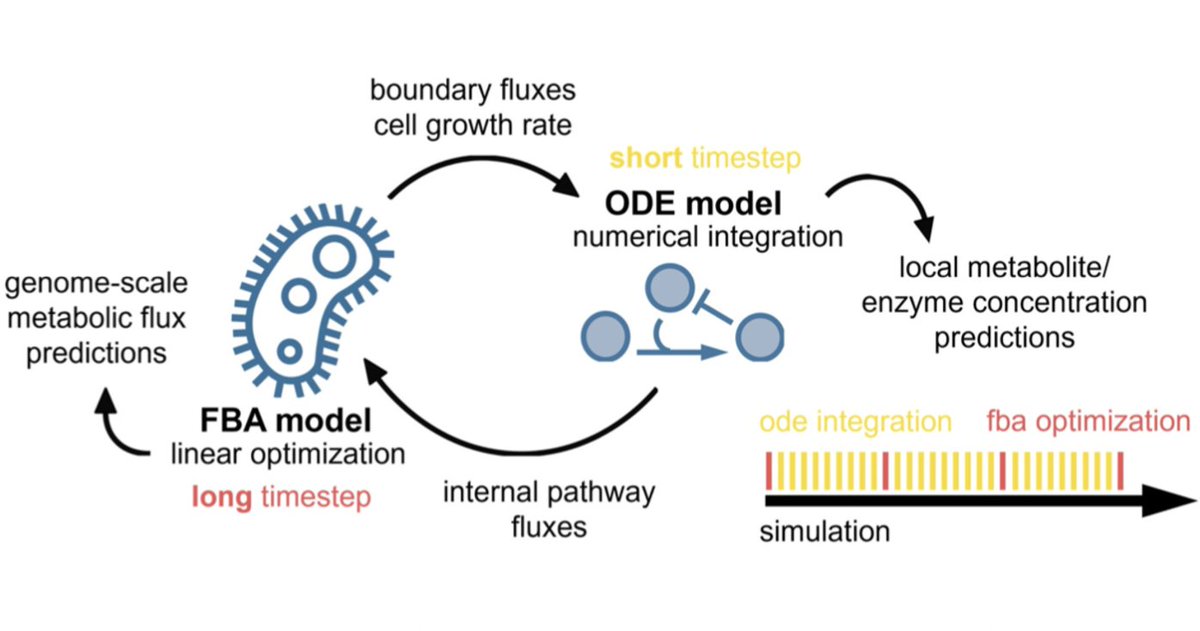
📢new preprint from our lab: Multiscale pathway simulation coupled with host metabolism. It combines genome-scale, kinetic and #MachineLearning models. biorxiv.org/content/10.110… Fab work by Charlotte Merzbacher with Oisin Mac Aodha #syntheticbiology #AI #Bioinformatics


Exhilarated to announce I've been tenured at DTU 🤯🥳. Thanks to Casey Greene and Tuuli Lappalainen for taking the time to evaluate my application 🙏 The timing is perfect, as I have 3 bioinformatic openings (both PhD and postdoc) - see👇 Retweets are sincerely appreciated 😊


I have several exciting openings for PhD students and postdocs in the context of my advanced ERC grant #ERCAdG on Assumption-lean (Causal) Modeling and Estimation. Please spread the news! users.ugent.be/~svsteela/ Ghent Data Analysis and Statistical Science @UGent_fwe Ghent University Research @UGent European Research Council (ERC)

📢New preprint from our lab Self-supervised learning for fluxomic data preserves structure across cell types Charlotte Merzbacher ANC@Edinburgh #sysbio #synbio #ML #metabolicmodelling #cobrapy biorxiv.org/content/10.110…