
Gonzalo Benegas
@gsbenegas
Postdoc @ UC Berkeley
ID: 531475019
https://gonzalobenegas.github.io/ 20-03-2012 17:37:12
82 Tweet
383 Followers
626 Following

Very glad to see our CherryML method published online in Nature Methods. The published version has updated and expanded results, so please check it out. doi.org/10.1038/s41592… (1/3)









We are delighted to share our work on a robust, accurate method for sampling genome-wide genealogies (aka ARGs) from the full posterior distribution. This is a product of years of hard work by Yun Deng 邓赟, who is jointly advised by Rasmus Nielsen and me. doi.org/10.1101/2024.0… 1/n

Highlighting some work I've enjoyed reading recently: "DNA language models are powerful predictors of genome-wide variant effects" by Gonzalo Benegas, Sanjit Batra, and Yun S. Song: pnas.org/doi/10.1073/pn… It's simply incredible unsupervised DNA language models can learn so much!




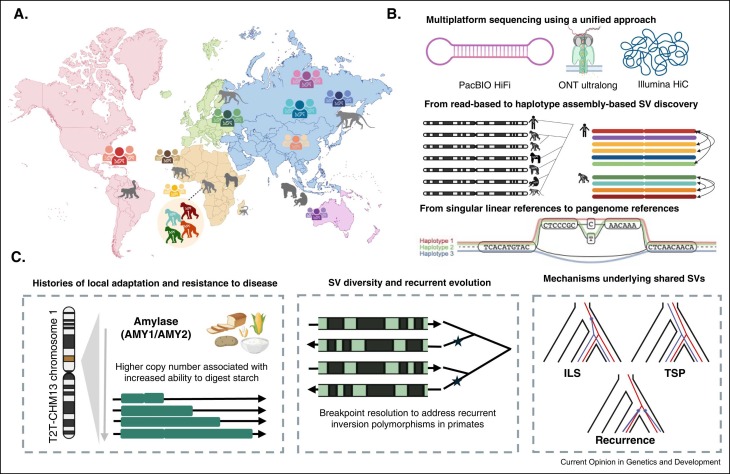
Had so much fun writing this small review with Nicolas Lou and Peter Sudmant about structural variation in primates in the era of T2T genomes and pangenomics :) sciencedirect.com/science/articl…

My main points at the #ICML2024 AI for Science panel (general guidelines, exceptions exist): -- Good projects are structured to have intermediate milestones that are measurable, thus creating constructive feedback loops. (I knew this one would get to Max Welling 😀) --

We are excited to share the preprint of the main project I worked on during my PhD: understanding the color polymorphism of the strawberry 🍓 🐸 poison frog in Bocas del Toro, Panama biorxiv.org/cgi/content/sh… Rasmus Nielsen


Our paper on the evolution of structural variation at the amylase locus is out today in Nature! nature.com/articles/s4158… congrats to co-first authors Joana L. Rocha Nicolas Lou Davide Bolognini Alessandro Raveane & Alma Halgren, co-corresponding author Erik Garrison




