Heidi Rehm
@heidirehm
Genomic medicine researcher; chief genomics officer at MGH; clinical lab medical director at @broadinstitute; Tweets/views are my own.
ID: 313939160
http://rehmlab.org 09-06-2011 13:14:44
825 Tweet
8,8K Followers
362 Following



We are delighted to announce this year's call for applications supporting postdoctoral fellows for the NHGRI-funded MGB T32 Training Program in Precision and Genomic Medicine cgm-dev.massgeneral.org/training-progr… Please RT or share w/ those who may be interested cc Jordan Smoller


We are delighted to publish guidance for applying segregation (PP1 & BS4) and phenotype specificity (PP4) evidence to variant classification - both important approaches to implicate the variant's locus in disease. Special thanks to Les and the whole team! authors.elsevier.com/sd/article/S00…


Dedicated, elegant detective work from Vijay Ganesh, CHASERR lncRNA Dad, and many others to pin down the mechanism whereby loss of one copy of a non-protein-coding RNA gene causes a genetic condition.


Progress and call to action for advancing access to genome sequencing for individuals and families with rare genetic disorders. Via npj Journals Genomic Medicine. nature.com/articles/s4152…


Today we announce the submission to #ClinVar of >32,000 variants evaluated for the return of hereditary disease risk in the AllofUsResearch cohort. Its wonderful to return value to those who contribute their data to biomedical research! More details here: allofus.nih.gov/news-events/an…

🎉 We are excited to announce our 12th Plenary keynote speakers: - Professor Dame Sue Hill, Transforming health and care - Phillip Wilcox, University of Otago - Alex Brown, telethonkids & the Australian National University - Herawati Sudoyo, PRBM Eijkman Register and learn more: bit.ly/3wfBFQ0





👥 GA4GH and the Research Data Alliance (Research Data Alliance (RDA)) have agreed to a Strategic Relationship to collaborate on areas of mutual interest and promote the advancement of responsible data sharing. Read the press release: hubs.ly/Q02Gk4xr0


There is still time to register for #GA4GH12thPlenary, co-hosted by Australian Genomics. Hear from Kathryn North, Heidi Rehm, Oliver Hofmann, and @Tiffboughtwood about why you should join us! 12th Plenary registration is available here: hubs.li/Q02JsDZK0

Estimating the probability of disease for people carrying a genetic variant - known as penetrance - is empowered by large population cohorts, but *very* easy to screw up. So Caroline Wright and friends (including me) explain how to do it right.


Wonderful news!!! Congrats Aaron Quinlan

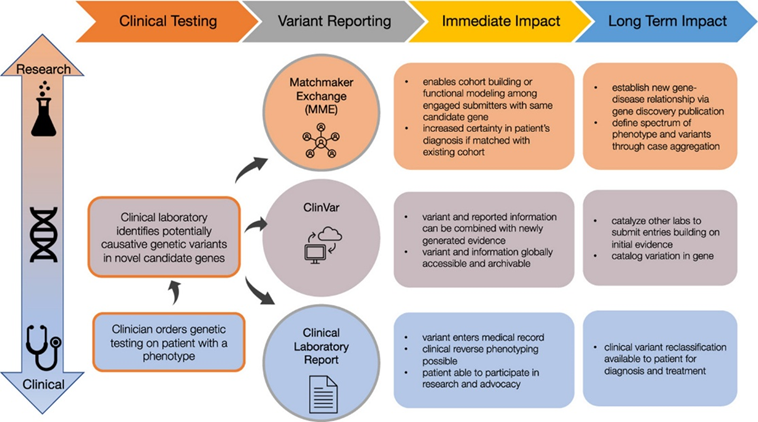
As part of GREGoR Consortium , we are excited to announce new guidance to encourage clinical labs to report novel candidate genes during clinical testing. We give criteria for triaging, sharing, and reporting novel genes through various methods. authors.elsevier.com/c/1jcDA3vlFV4E…