
Maurice Lee
@maurice_y_lee
I pipette, build microscopes, & write code | he/him |
Biophysics PhD |
Developing 3D spatial transcriptomics (MERFISH) in khchenlab.github.io @astar_gis
ID: 4710391092
https://scholar.google.com/citations?user=BYjdgGUAAAAJ&hl=en 04-01-2016 21:29:39
11,11K Tweet
5,5K Followers
2,2K Following




We've been asked (thanks Stan Schwartz!) for a summary of this discussion, which I think would benefit people who couldn't attend! I didn't take notes, but I will share from my memory and warmly invite other panelists and #SMLMS2024 attendees to add to the thread!


A paper in Nature Machine Intelligence discusses a deep learning method that recognizes specific nuclear signatures, which can identify cellular heterogeneity and differentiate between various cell states using a small amount of super-resolution microscopy data. go.nature.com/3MyyqaR



Out at Nature Methods just now! nature.com/articles/s4159… We present #SN2N, a flagship level unsupervised denoising solution. It’s fully competitive with supervised learning without needing large training-set and clean ground-truth, using 1 noisy frame for training. (1/n)
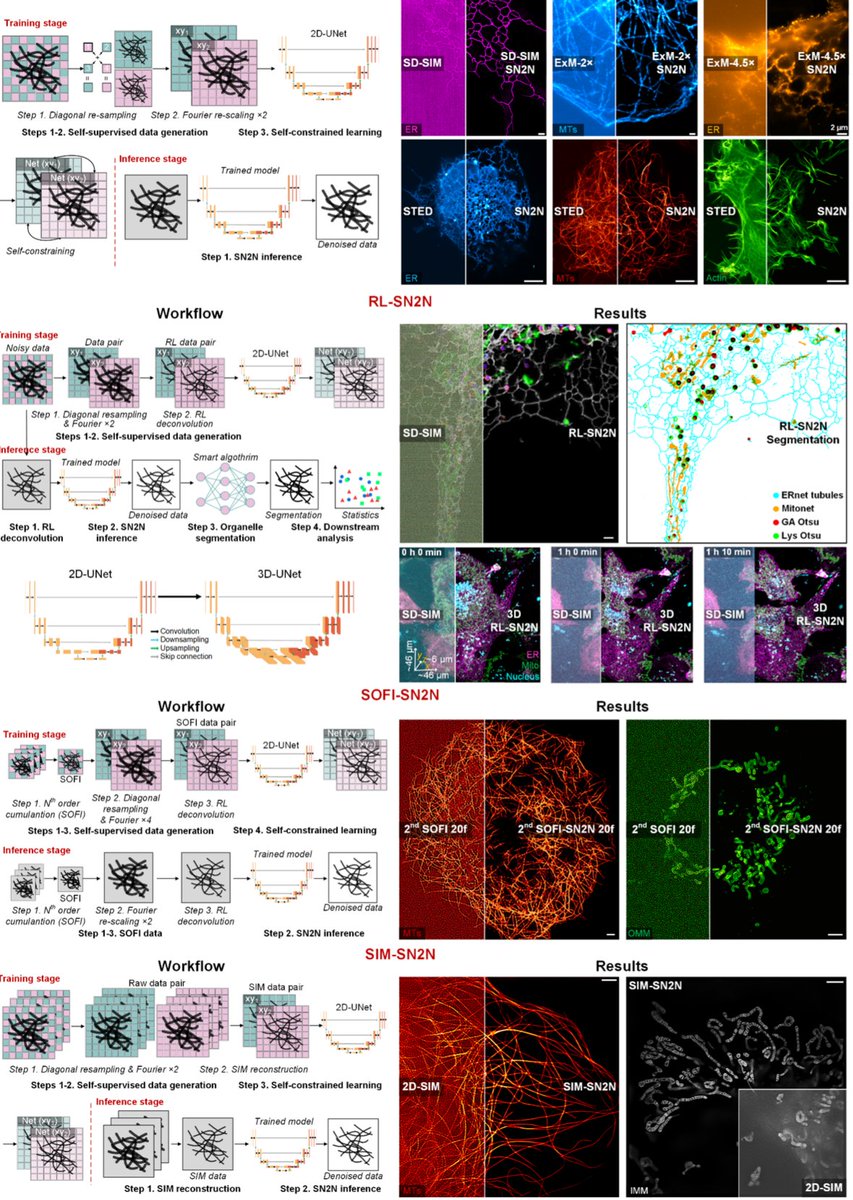

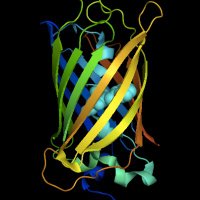

We developed a new super-stable RFP for advanced microscopy, like CLEM, Rapid tissue clearing, Live 3D-SIM and live STED. Read this #preprint on Research Square: mYongHong: a structure-guided design of stable and monomeric red fluorescent protein researchsquare.com/article/rs-471…




