
Matteo Gentili
@mgentili_
🇮🇹PostDoctoral Fellow, The Broad Institute of MIT and Harvard
ID: 107422534
https://mstdn.science/@mgentili 22-01-2010 14:18:41
1,1K Tweet
434 Followers
434 Following


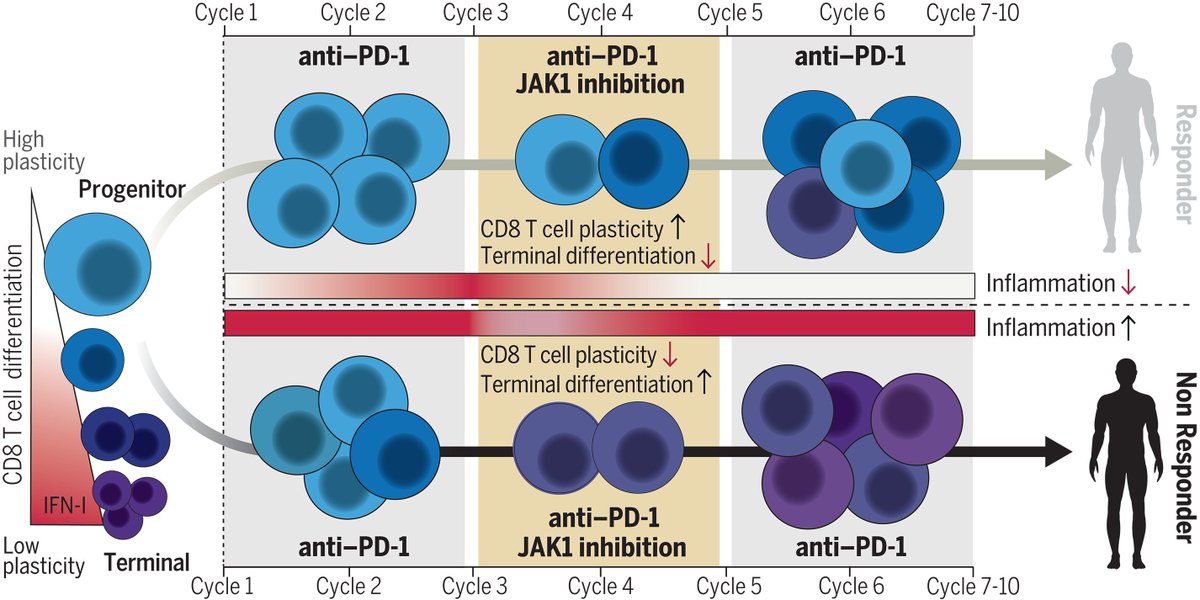
Janus kinase (JAK) inhibitors improve antitumor responses Science Magazine Melina Marmarelis Andy Minn E. John Wherry Veronika Bachanova MD Teijaro Lab science.org/doi/10.1126/sc… science.org/doi/10.1126/sc… science.org/doi/10.1126/sc…



REVIEW Nature Immunology The balance of STING signaling orchestrates immunity in cancer nature.org/articles/s4159…




Latest from us. How a rare genetic disorder inspired a broad spectrum antiviral drug. Started by Justin Taft continued by Roosheel Patel and Dr. Akalu in our lab. Columbia Children's Health Lipschultz Precision Immunology Institute Icahn School of Medicine at Mount Sinai Columbia University nam02.safelinks.protection.outlook.com/?url=https%3A%…

VEXAS‐Defining UBA1 Somatic Variants in 245,368 Diverse Individuals in the NIH All Of Us Cohort - 2024 “Variant allele fractions were lower than reported in clinical VEXAS cohorts, and bioinformatic analysis detected no clinical manifestations of VEXAS.” acrjournals.onlinelibrary.wiley.com/doi/10.1002/ar…

Combining IFNAR inhibition with mRNA-LNP vaccination promoted specific stem cell-like memory T differentiation and enhanced protection against chronic infection Joanna Groom The Pardi Lab biorxiv.org/content/10.110…


For 25 years, the Toll-like Receptor pathway was considered a series of distinct protein complexes that drive inflammation. Today, we report that the entire pathway, from receptor to transcription factor, is executed from within one complex—the myddosome. nature.com/articles/s4158…

#InterferonPower! All re type I IFNs Science Immunology by CrowLab & Casanova Lab: “only a narrow window of IFN-I activity is beneficial”! Insufficient IFN-I drives life-threatening viral diseases while excessive IFN-I causes inflammatory interferonopathies bit.ly/SciImm_adm8185

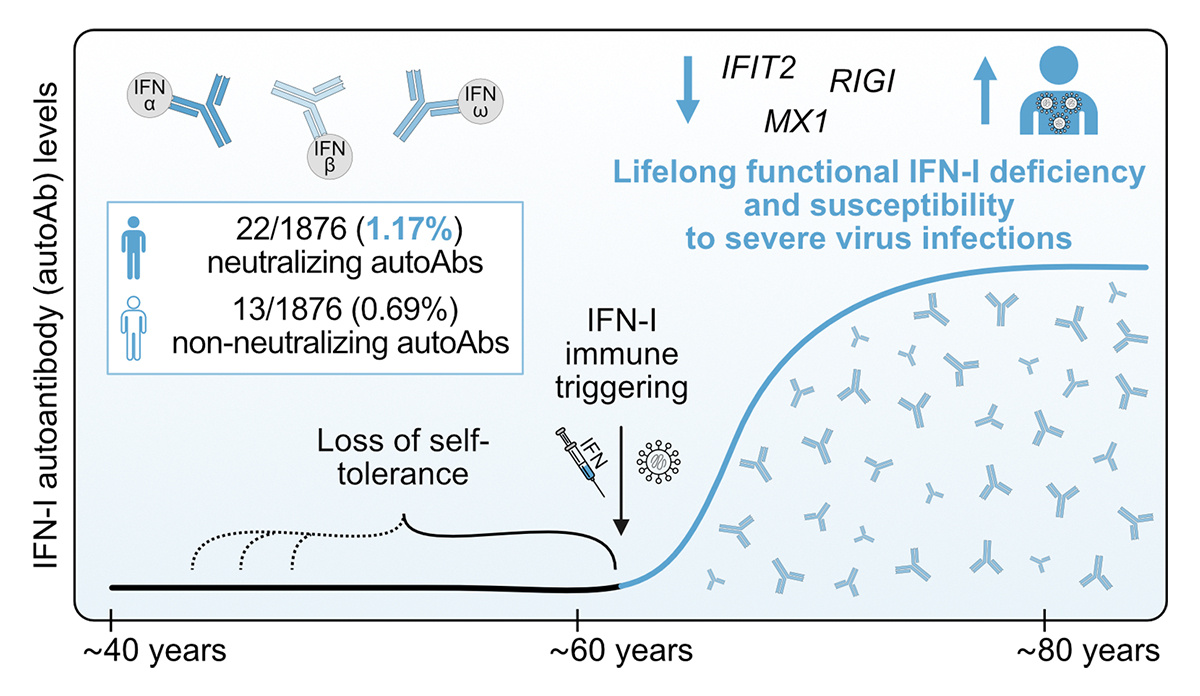
.Sonja Fernbach, Mair, Ben Hale UZH Medical Virology and colleagues dissect how rare anti-IFN-I autoantibodies develop and weaken defenses over time, making some individuals more susceptible to infections. hubs.la/Q02H58D10 #Autoimmunity #Immunodeficiency #InfectiousDisease





Thrilled to share our new paper in Science Magazine, in which we identify p62/SQSTM1 as a critical regulator of micronuclei 🧬🔬🧫 #ScienceResearch science.org/doi/10.1126/sc…


