Rushin Gindra
@rushin_gindra
PhD candidate @SaurLab at TranslaTUM and @helmholtz_ai, developing deep learning methods for Spatial Transcriptomics
ID: 816643046041231360
04-01-2017 13:50:49
51 Tweet
155 Followers
195 Following



Excited to share our latest work: Combining #graphs and #transformers for robust weakly #supervised learning for whole slide image analysis. Published IEEE-TMI (doi.org/10.1109/TMI.20…) with YZ, Kolachalama laboratory, Jennifer Beane BU CompBioMed, Boston U Chobanian & Avedisian School of Medicine, BU AIR, Hariri Institute for Computing, Boston University




Another glorious preprint from the Johns Hopkins Kimmel Cancer Center Convergence team (Dr. Luciane T Kagohara 🇧🇷🧬👩🏻🔬👩🏻💻 et al): Spatial transcriptomics of FFPE pancreatic intraepithelial neoplasias reveals cellular and molecular alterations of progression to #PancreaticCancer biorxiv.org/content/10.110…




As PhD application deadlines approach, we are super excited to announce cs-sop.org, created by Zhaofeng Wu Alexis Ross @ZejiangS💻 cs-sop.org is a platform with statements of purpose generously shared by previous applicants to CS PhD programs 🧵(1/n)






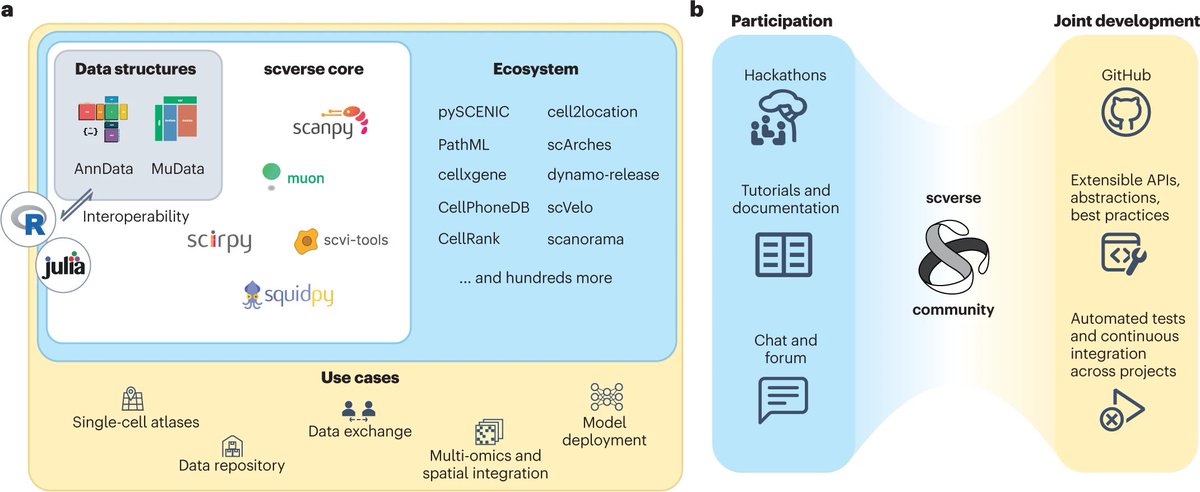
Glad to see our review on best practices for single-cell analysis across modalities out Nature Reviews Genetics! In a big team effort led by Lukas Heumos & Anna Schaar, we recommend workflows based on benchmarks. Paper at rdcu.be/c8RU5 & extension at sc-best-practices.org.