Sangjin Kim
@sjkimlab
Studying how molecules work together in vitro, in vivo, and in silico. Biophysics research group @ University of Illinois, Urbana-Champaign
ID: 1108576626533715968
https://www.sjkimlab.org/ 21-03-2019 03:50:38
398 Tweet
494 Followers
391 Following

We started out making dyes for single-particle tracking in cultured cells. But now we have dyes that enable pulse-chase labeling in vivo. Read the preprint here, a project driven by the indefatigable Boaz Mohar (@[email protected]) with other colleagues at HHMI | Janelia biorxiv.org/content/10.110…

Our new review is out in Nature Reviews Microbiology: Molecular Mechanisms of Antibiotic Resistance Revisited. Dr Lizzy Darby Ilyas Alav Mark Webber Eleftheria Trampari Maria Solsona & Pauline IMIBirmingham Quadram Institute #WAAW 😃 nature.com/articles/s4157…



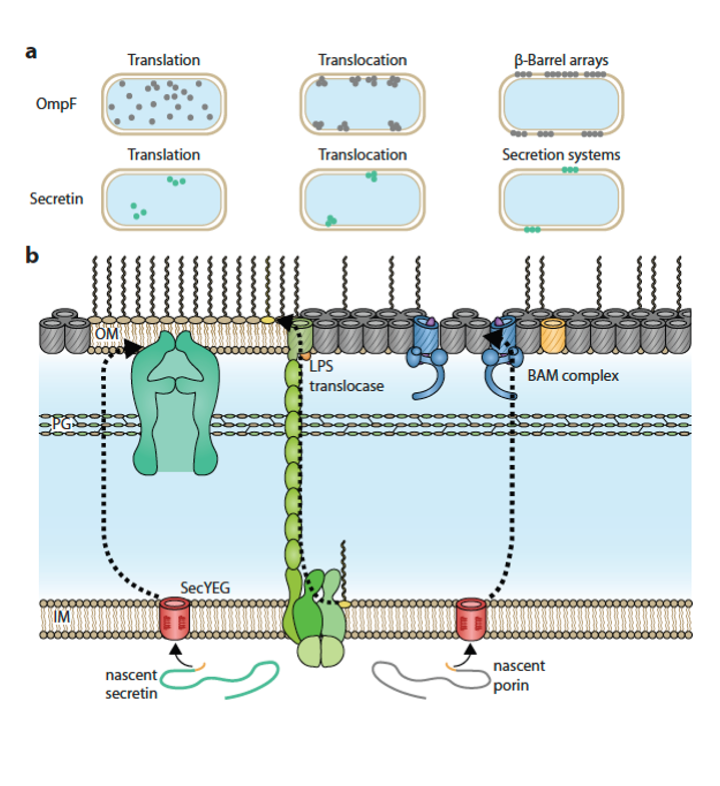
It was impressive reviewing the papers from our field to show how far understanding has come for the quantitative aspects of bacterial cell surfaces and the processes by which they are made rdcu.be/c6iIS Centre to Impact AMR MACSYS-CoE Monash Biomedicine Discovery Institute


Thermal agitation is slow and ineffective when it comes to self-assembling soft materials. Can active baths do better? Today in Nature Physics, we show that a (chiral) active bath can rapidly⚡️build extraordinary structures unreachable to thermal physics👽 nature.com/articles/s4156…

RNA polymerase generates equal amount of positive and negative supercoils upon DNA unwinding, and does so in an RNA independent manner. Intriguing dtudy from CD lab. Very interesting! cees dekker biorxiv.org/content/10.110…

Thank you Katsu Murakami for inviting me! I had a great day talking with your faculty and students!


A dream of our lab has been to image the full central dogma from a single endogenous gene, all live and with single molecule resolution. After many years we are happy to unveil a beautiful cell line that makes it possible. Check out our preprint (biorxiv.org/content/10.110…)! (1/n)



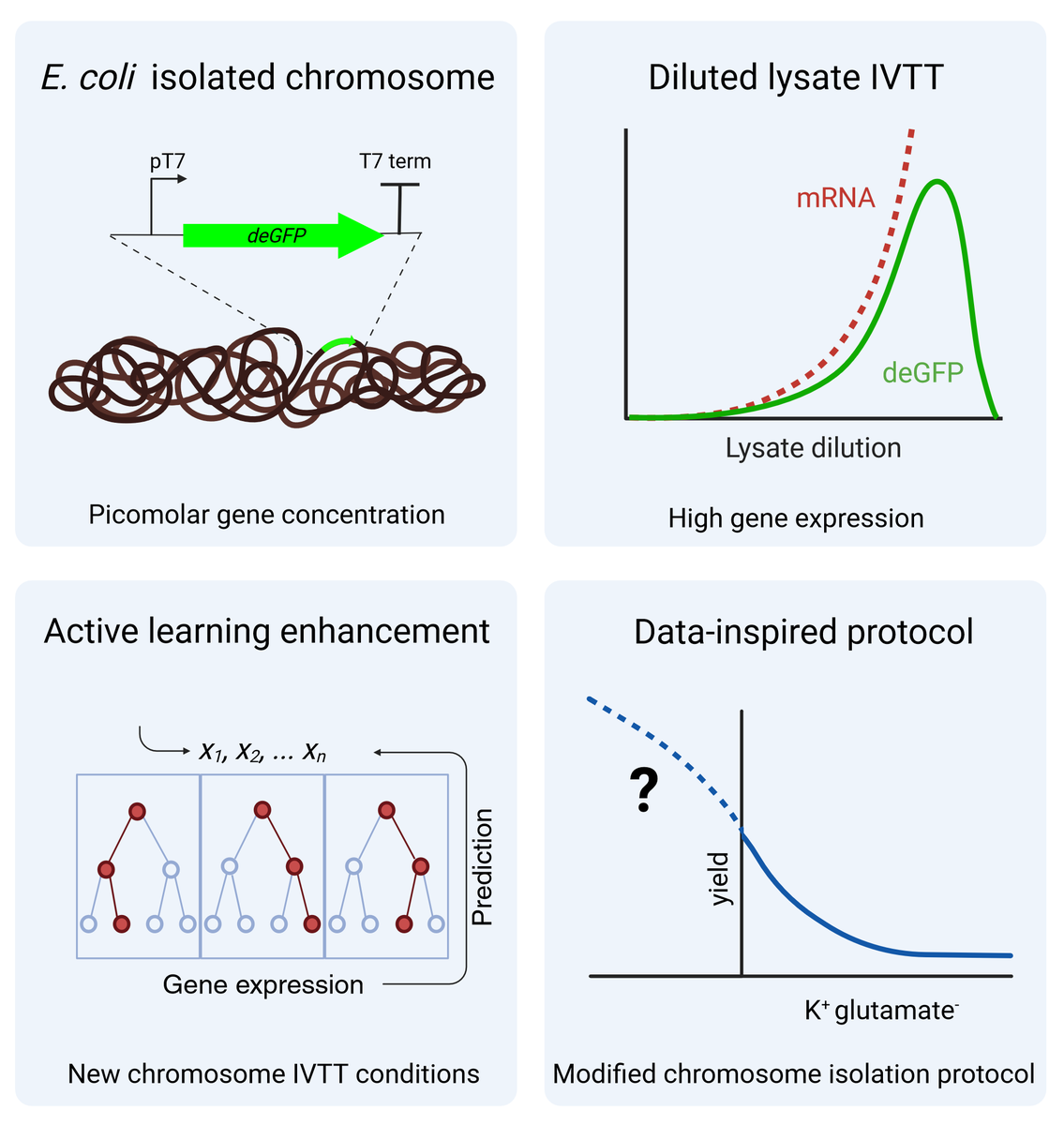
For building a synthetic cell, we will need to access the information carried in large DNA molecules. We used active learning to adapt in vitro gene expression to E. coli's chromosomes with a surprising efficiency. Synthetic Cell initiative Now out in ACS_Omega 🥳 👉doi.org/10.1021/acsome…

Yay! Kim lab finished strong in the Illinois 5k race Illinois Marathon

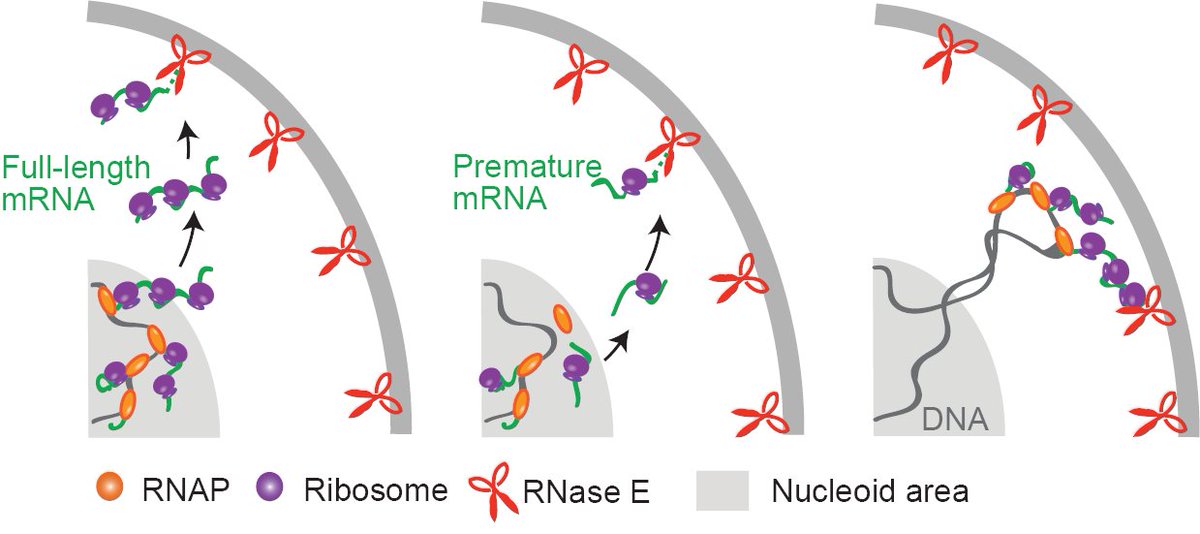
So excited to announce our new method for multiplexed RNA imaging in bacterial cells: bacterial-MERFISH! A huge congratulations to the team of Ari Sarfatis, Yuanyou Wang, and Nana Twumasi-Ankrah! Check out the following thread or our bioRxiv (biorxiv.org/content/10.110…) 1/12


Check out our latest findings on the epitranscriptome of Escherichia coli biorxiv.org/cgi/content/sh… , spearheaded by #Sebastian Riquelme-Barrios and in excellent collaboration with Pascal Giehr 🕊🇺🇦 and Kaiser Lab; funded by @sfb1309.