
Evan Newell
@evnewell1
Trying to understand human immunity by looking through the eyes of antigen-specific T cells.
Lab at Fred Hutchinson, Vaccine and Infectious Disease Division
ID: 911591183733760005
https://research.fhcrc.org/newell/en.html 23-09-2017 14:00:49
902 Tweet
1,1K Followers
982 Following

Terrific work by Daniel C. Jones in Evan Newell lab- Proseg is our go-to cell segmentation for all spatial transcriptomics data!
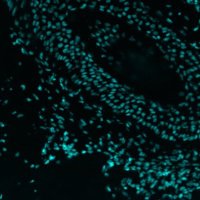
FHIL says check out Proseg cell segmentation by Daniel C. Jones for spatial transcriptomic projects. We use it for our Xenium & CosMx data and it rocks!🎇 biorxiv.org/content/10.110…

RECOMMENDED! Our initial tests were like…wait what? 🤯 I’m sure Fred Hutch Innovation Lab Evan Newell would love some feedback. Cell Simulation as Cell Segmentation | bioRxiv biorxiv.org/content/10.110…
Luciano Martelotto 🛠🧬💻🇦🇺 Fool Fred Hutch Innovation Lab Evan Newell Quick analyses in our lab also show it performs really well. We'll show some plots soon.

#FixNCut, a method for early sample preservation to overcome current limitations in Single-cell omics | CNAG Adelaide Centre for Epigenetics Uni of Adelaide St. Jude Research Omniscope South Australian immunoGENomics Cancer Institute Peter Mac Cancer Centre cnag.eu/news/fixncut-n…



1🔬/ Excited to share our latest preprint led by Shuxiao Chen , Shenggao , Garry P. Nolan , Sizun Jiang , Zongming Ma , and with help from Shalek Lab, Scott and more! biorxiv.org/content/10.110…


Wanna annotate cells in your spatial-omics data but tired of just calling them by their general phenotypes? Hey we got the solution for you! See awesome thread by the hotshot Bokai Zhu, and working with maestros Zongming Ma and Garry P. Nolan

Our former collaborator Evan Newell + others explore ways to determine #cell segmentation accurately in #spatial #transcriptomics in this preprint. bioRxiv Fred Hutchinson Cancer Center pubmed.ncbi.nlm.nih.gov/38712065/


Our first Parse Biosciences Evercode TCR experiment- big appeal is banking fixed cells for TCR. We benchmarked sorted T-cells from 4 healthy donor PBMCs, fixed with v2 reagents, and loaded slightly over the recommended 350k cells to aim for 100k, generating 8 WT & TCR sublibraries. (1/7)



Very excited to have Mellon out in Nature Methods today!




Earlier this week we carried out a preliminary analysis of our first 10x Genomics Xenium prime (5,000 gene) run. We had designed this experiment to get a sense of how the prime chemistry performed compared to the Xenium V1 chemistry.

What did we #learn after #sequencing #5million #Tcells #scRNA and #TCRseq with Lisa Wagar Andreas Tiffeau-Mayer Suhas Sureshchandra and funding agency Wellcome Leap STLChildrensHospital -Tissue determinants of the human T cell receptor repertoire. biorxiv.org/content/10.110…


