Jue Wang
@jueseph
Research scientist @Deepmind. Formerly @UWproteindesign @ginkgo @HMS_SysBio.
ID:25824880
http://juewang.strikingly.com 22-03-2009 14:43:16
2,9K Tweets
1,5K Followers
782 Following
Follow People

Just discovered Grant Sanderson videos and am now obsessed. Some examples: youtube.com/watch?v=pQa_tW… youtube.com/watch?v=X8jsij…
Join us in 1 week to hear Brian L Trippe and Jason Yim talk about diffusion models of protein backbones!
As always, sign up here for updates and zoom links: ml4proteinengineering.com



Awesome work! I made a Sergey Ovchinnikov 🇺🇦 -style Colab notebook for inpainting loops. Hope people find it useful! colab.research.google.com/github/polizzi…



Like a word processor’s autocomplete function, AI designed Institute for Protein Design draws on its understanding of how proteins fold to fill in additional parts of the protein around the central feature. bit.ly/3OEqfZc Jue Wang Ian Haydon




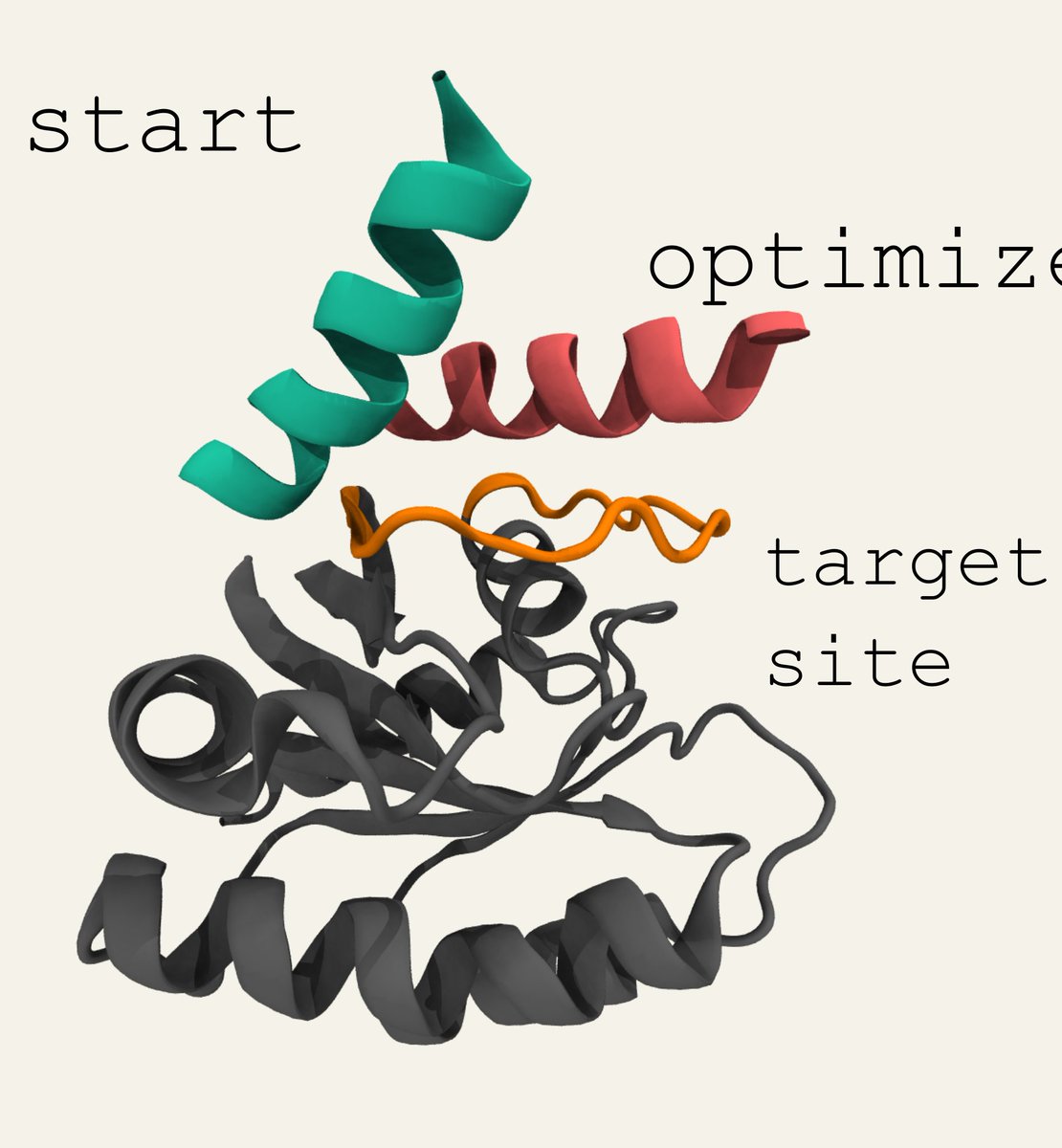
One big leap between preprint and publication is that the inpainting method became very powerful and is now the method of choice for motif-scaffolding problems. Along with ProteinMPNN and AF2-based filtering, it has completely changed protein design at Institute for Protein Design. 8/8

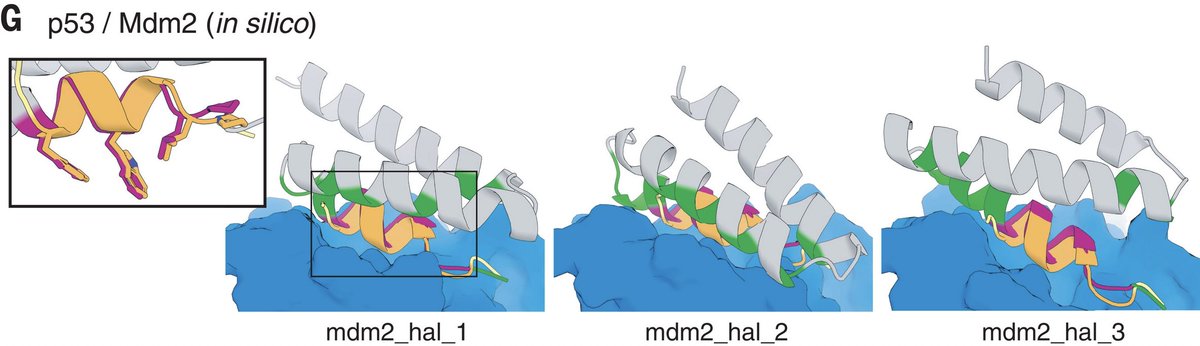
Sidney Lisanza and I used hallucination to create protein binders, both by scaffolding a binding motif, and with only the target as input. This 'de novo' binder hallucination uses the ability of RosettaFold to model interactions between protein chains. 7/8

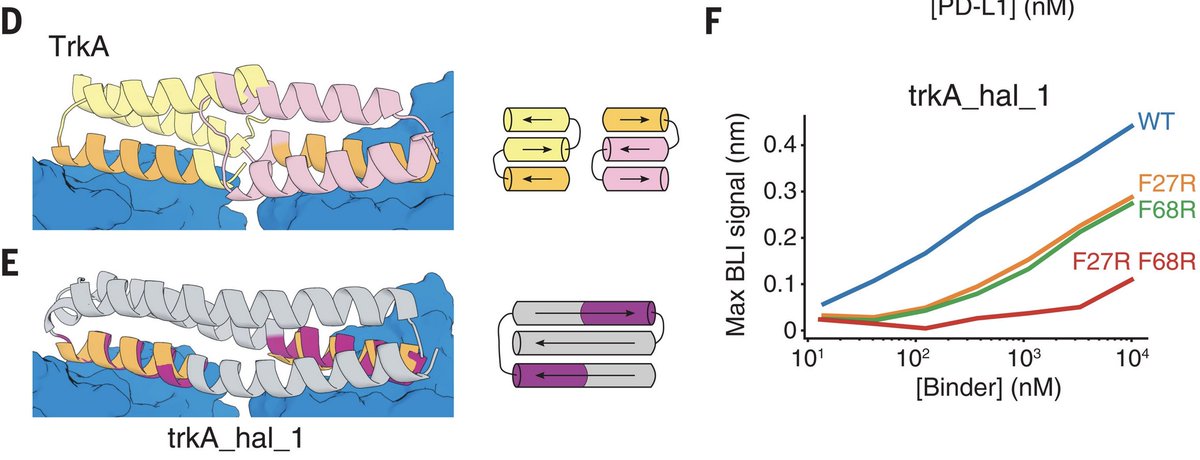
Doug Tischer linked together 2 binding sites to create a symmetric dimeric binder of the TrkA receptor involved in various diseases, showing that we can design binders to multiple targets in a geometrically defined way. This is crucial for modulating real signaling pathways. 6/8

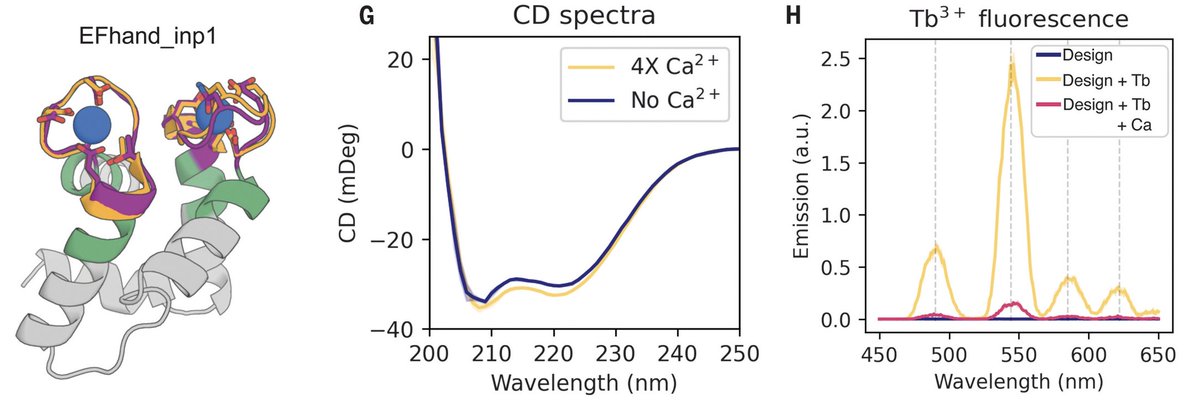
We also successfully designed and tested metal- and protein-binding proteins. This shows the power of the inpainting method developed by Joseph Watson and David Juergens: from inputs with short helices or loops, we got outputs of well-packed, ideal helical bundles. 5/8