
Oxford Nanopore
@nanopore
From portable to population scale, nanopore devices offer direct analysis of DNA/RNA in-real time — towards the analysis of anything by anyone, anywhere.
ID:37732219
http://www.nanoporetech.com 04-05-2009 19:21:00
19,4K Tweets
65,2K Followers
9,9K Following
Follow People


Actionable methylation detection and CNV calling from low pass WGS to enable discrimination of tumor types from 210Mb of data (Oxford Nanopore MinION scale). From FFPE sample.
One less barrier. Thanks to pioneering work of Christian Thomas et al.




One of our customers, Seoul University, tested methylation using the RNA004 kit with early access. The results are quite amazing: improved accuracy and output. We are looking forward to the upcoming release of RNA004 Oxford Nanopore.


Amazing to see Maximilian Gantz and team using MinION in the Peruvian Amazon to study the gut microbiome of the ocelot!
A fantastic example of how portable, real-time DNA/RNA sequencing can drive impactful ecological studies.
#AnythingAnyoneAnywhere #10YearsON

Excited to be at #ECCMID2024 #ESCMIDGlobal representing the Oxford Nanopore #EPI2ME team with Natalia Garcia - We'll be demoing AmPORE-TB, Metagenomics, and Isolate (NO-MISS) every day at 12:15pm on our booth ( #C3 )! Be sure to join us!

Don’t miss Oxford Nanopore exhibiting at The Festival of Genomics & Biodata in Boston in June! Grab your ticket here: hubs.la/Q02ty0Hl0 #FOGBoston



Have a read on my new piece on #biomarker discovery in #oncology for #pharma and #biotech in
rdworldonline.com/for-drug-disco…
Note the value of Oxford Nanopore flexible platform: short and long reads as well epigenetic information included.
What you are missing matters!

AmPORE-TB is a same day (<6hrs) assay for the identification of Mycobacterium tuberculosis and antimicrobial resistance markers.
Join us during one of our demos at #ECCMID2024 to learn about this new end-to-end workflow coming later this year: bit.ly/3Q40TaI


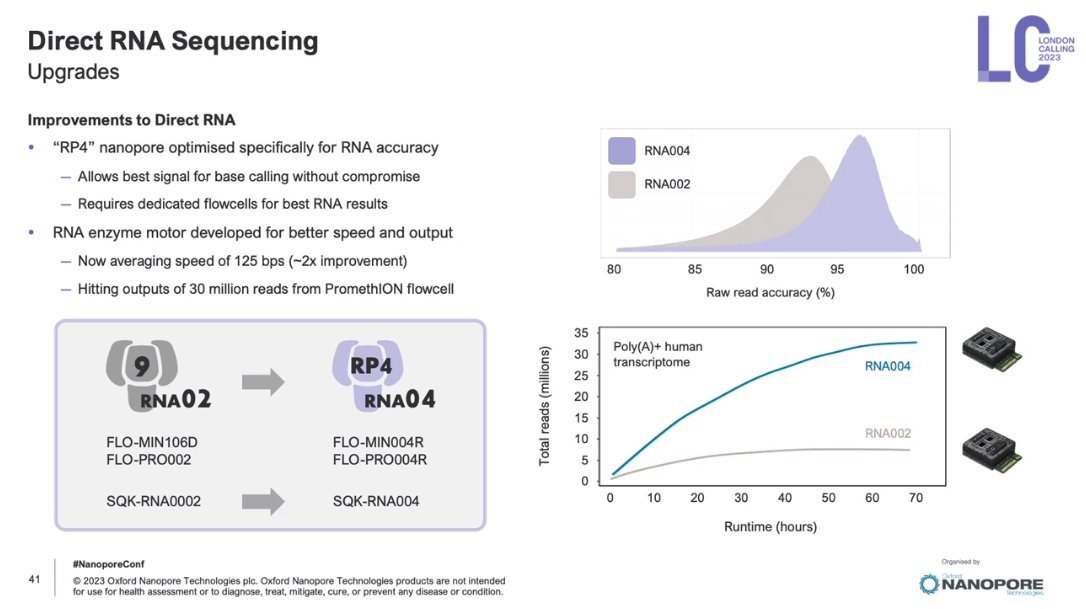
Drastic improvement for Oxford Nanopore DRS using kits RNA004 vs RNA002, confirming a similar observation previously by other labs Alexander Wittenberg Miten Jain. Average base error rate reduced from 10.2% (RNA002) to 4.2% (RNA004).



From an initial sketch on the side of the road, to the only sequencing technology that provides native, real-time analysis of DNA & RNA fragments of any length, in fully scalable formats. Nanopore sequencing has come a long way...
Learn more: bit.ly/3UgKXV5 #TechTuesday












