Heng Li
@lh3lh3
Associate professor of biomedical informatics at @HarvardDBMI and @dfcidatascience
ID:2165843324
http://liheng.org 31-10-2013 02:21:20
3,7K Tweets
10,0K Followers
208 Following
Follow People


We have released some raw Oxford Nanopore data for benchmarking purposes, along with some basecalled outputs.
We use the subset of 20k or 500k for benchmarking tools and methods a LOT!
We thought they would be helpful to others too.
4/5kHz R10.4.1 & RNA004
gentechgp.github.io/gtgseq/







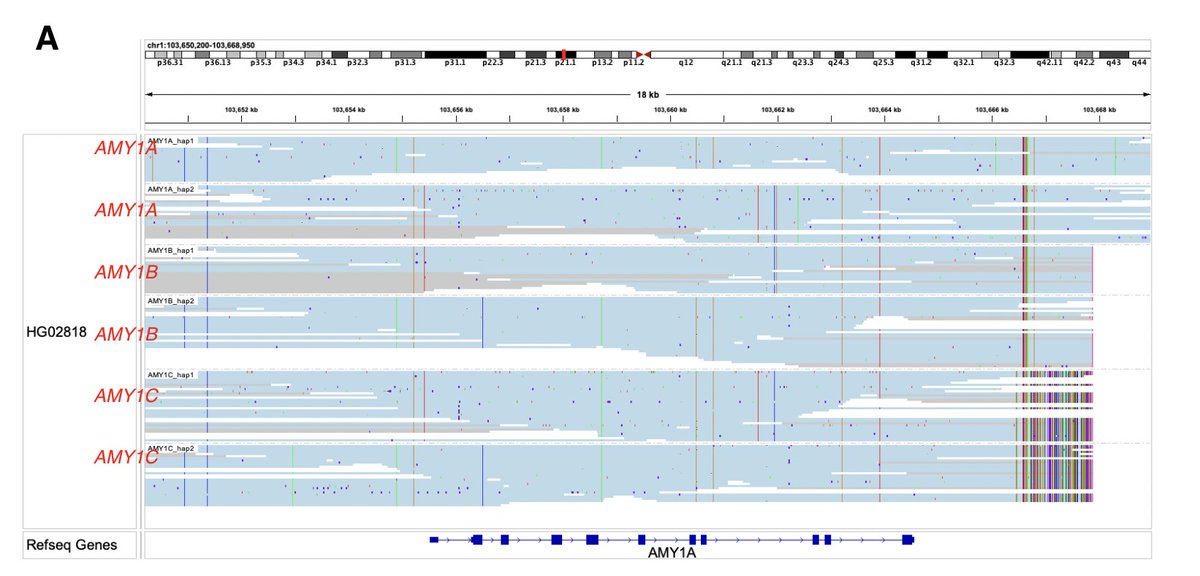
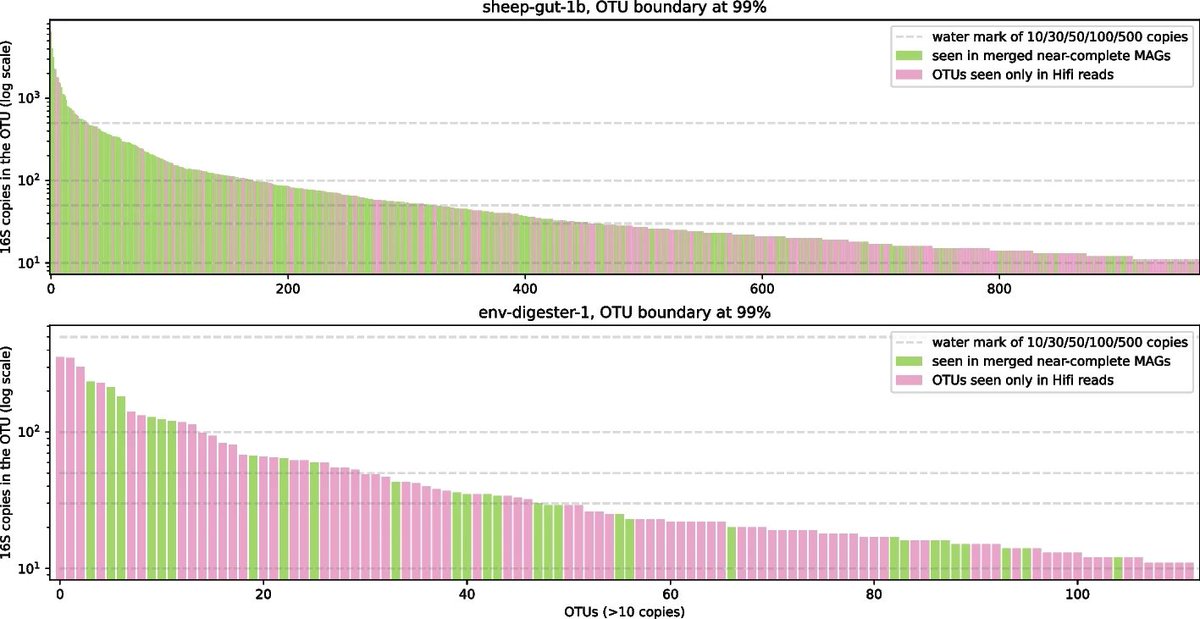
Xiaowen's (Xiaowen Feng) new paper uses 16S in reads and k-mers to answer how much a metagenome assembly captures the species and sequence content in a sample. We found abundant species are not always assembled and proposed a heuristic to rescue some of them. genomebiology.biomedcentral.com/articles/10.11…


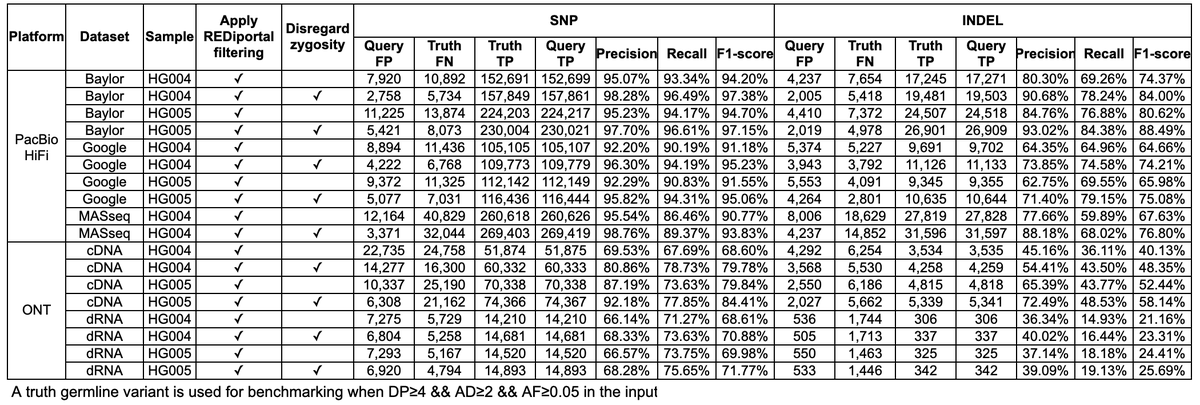
Making Clair3-RNA available, which is a side project of my lab inspired by Fritz Sedlazeck and Miten Jain last December. It's for RNA long-read germline variant calling. It supports ONT r9 cDNA, ONT rna002 dRNA, PB Sequel2 Iso-Seq, PB Revio MAS-Seq. 🔗github.com/HKU-BAL/Clair3… 1/n




Analysis of the limited M. tuberculosis accessory genome reveals potential pitfalls of pan-genome analysis approaches biorxiv.org/cgi/content/sh… #biorxiv_bioinfo


🌟1/3: Introducing mm2-gb, GPU-accelerated Minimap2 for long-read DNA mapping!
🔥 mm2-gb accelerates Minimap2's bottleneck (chaining) on GPUs without compromising accuracy. 🚀 Kudos to Xueshen Liu Joy Dong, Satish Narayanasamy & Gina Sitaraman Computer Science and Engineering at Michigan AMD Michigan Engineering