Ryan Wick
@rrwick
Bioinformatician at @TheDohertyInst
ID: 1677127723
http://github.com/rrwick/ 17-08-2013 02:08:23
988 Tweet
3,3K Followers
415 Following

Ryan Wick, myself and the team (Louise Judd Tim Stinear Rob Edwards & Sarah) have recently looked at the effect of depth on short-read polishing almost-perfect Nanopore bacterial genomes biorxiv.org/content/10.110… (1/7)


Excellent talk by George Bouras on short/long/hybrid assembly strategies for optimum chromosome and plasmid assemblies! Loads of information, advice, and great new #bioinformatics tools to use next. #ECCMID24


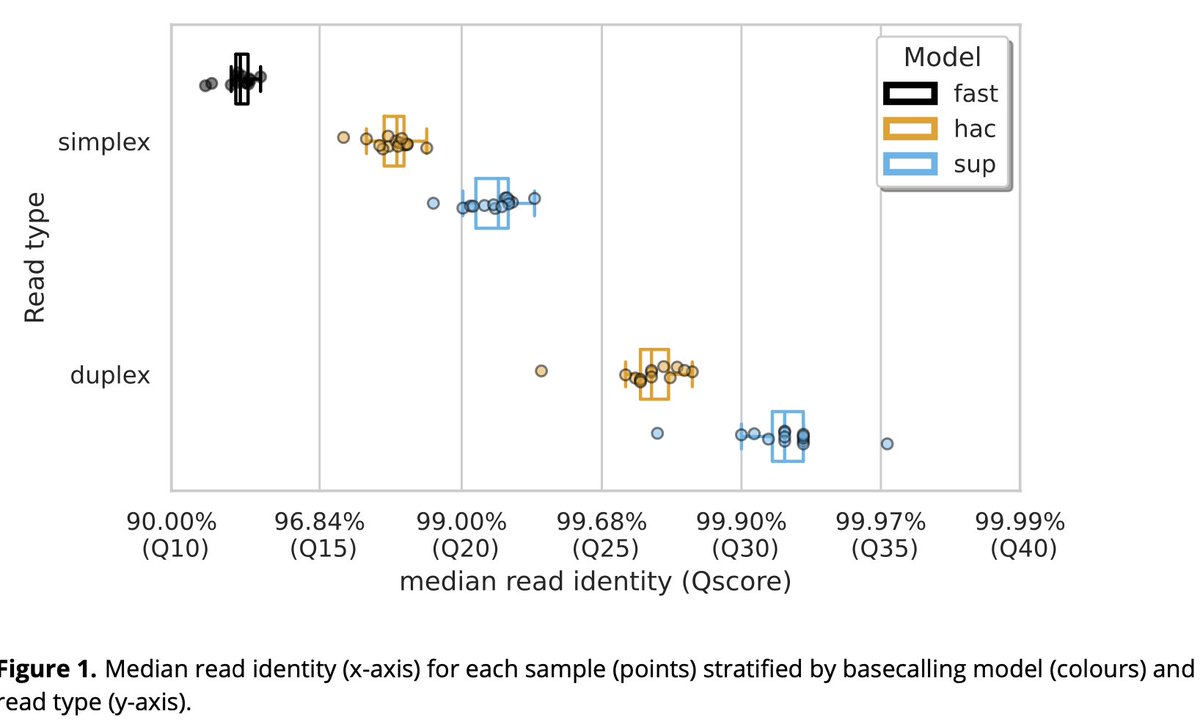
Is Oxford Nanopore duplex basecalling beneficial for bacterial genome assembly? I did some tests, and it seems like it might be! New blog post here: rrwick.github.io/2024/05/08/dup…

Are you a fan of #klebsiella genomics? Looking for a post-doc role? ...🥁 ... I'm recruiting! Join my team Translational Medicine at Monash University to help us build the first Klebsiella clone risk framework. See details here ⬇️ tinyurl.com/yrka232h Applications close June 9th. DM qs #klebNET Klebsiella Club

I'm recruiting for a new post-doc role based in Melbourne and contributing to #klebNET-GSP activities on #klebsiella genomic surveillance. Sylvain Brisse Kat Holt Centre for Genomic Pathogen Surveillance Chiara Crestani Tom Stanton (he/him) Sophia David Theycallmemaggie Check out the details ⬇️



The final version of our short-read polishing manuscript (headed by Ryan Wick of course) is out in Microbiology Society MGen! Tldr: with R10.4.1+Dorado ONT assemblies, few errors remain, so short-read polishing can make assemblies worse at low depth; target >=25x doi.org/10.1099/mgen.0…




I finally got around to testing HERRO (github.com/lbcb-sci/herro), the new Oxford Nanopore read correction algorithm which takes simplex ONT reads to PacBio-HiFi-level accuracy. I was impressed! Findings are on my blog in a two-part post: rrwick.github.io/2024/07/26/her… rrwick.github.io/2024/07/31/her…